Datasets
We overview the datasets released through the FALCON challenge. Technical details and the dataset itself are provided in the linked DANDI repository and the associated paper. In FALCON, we release a collection of five datasets spanning a variety of tasks, subjects, and brain areas. Though in our benchmark we are only evaluating a specific set of metrics for each dataset, we encourage the use of these datasets beyond the scope of our current competition.
H1
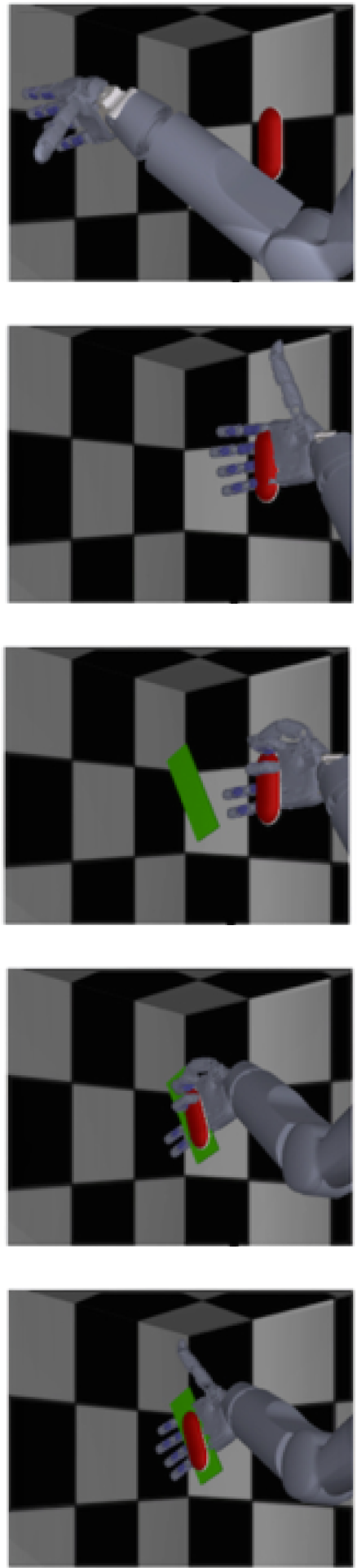
H1 contains open-loop calibration data collected in one human participant for a reach and grasp BCI experiment. The participant watches a virtual arm perform a series of movements and attempts to match these movements. The participant cannot execute these movements fully due to high level spinal cord injury, and for these datasets, their native limbs were completely at rest. The virtual arm behavior is phasic and pre-specified, such that movement are split into translation, orientation, and grasp phases. Neural activity is collected from Utah arrays implanted in the arm and hand areas of motor cortex. This data is provided by the Rehab Neural Engineering Labs at University of Pittsburgh.
Why H1?
Motor population activity represent a breadth of movement variables that are accessible through linear decoding. This richness has been used in microelectrode-based BCIs to enable high-dimensional arm and hand control first demonstrated over a decade ago. The saliency of movement-related information has been a hallmark motivation for microelectrode-based BCIs. Nonetheless, two challenges remain:
High dimensional control remains a burden to calibrate, with this particular experimental protocol demanding several minutes of calibration.
The identified decoders do not provide stable control, such that experimenters routinely calibrate new decoders in each experimental session despite the burden.
Providing methods to improve the efficiency of calibration in novel sessions would thus greatly improve the practical viability of high dimensional neuroprosthetic motor control from spikes.
H2
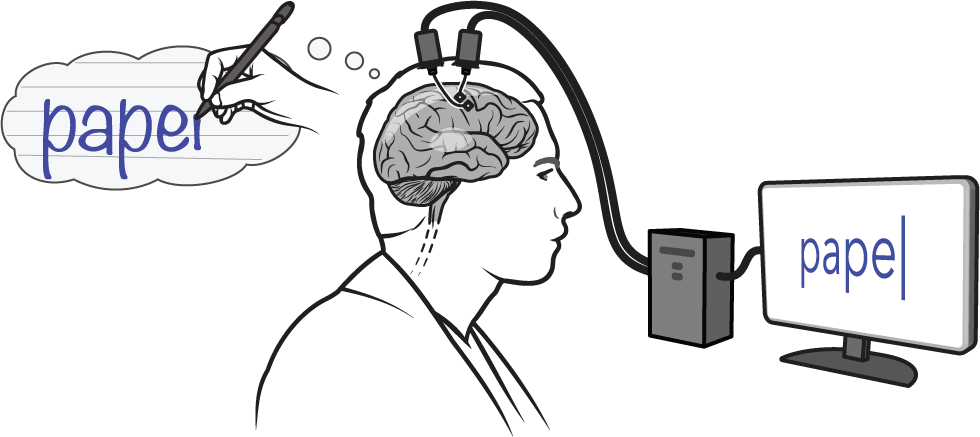
H2 contains data from a human iBCI partipant performing a handwriting task. On each trial, the participant is given a cue to copy by attempting to write each letter in the prompt. Neural activity is collected from two Utah arrays implanted in the hand “knob” area of cortex. This data is provided by Chaofei Fan at Stanford University.
Why H2?
The H2 dataset falls in the domain of brain-to-text BCI, which aim to restore communication capabilities. Decoders for this task will need to be able to accurately predict the intended character as well as determine when that character was intended to be written, as the task is fully self-paced. This task is therefore not amenable to traditional linear decoders. Additionally, decoding may make use of large language models, allowing a new class of recalibration approaches.
M1
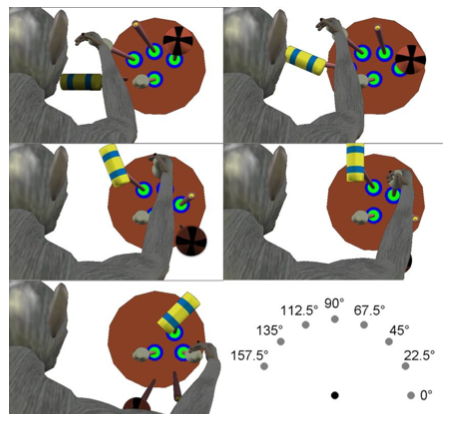
M1 contains data from a monkey performing a center-out reach-and-grasp task. The dataset comprises neural activity recordings from a rhesus monkey equipped with 6 floating microelectrode arrays, each with 16 channels. Additionally, intramuscular electromyography (EMG) data is captured from 16 locations in the monkey's right hand and upper extremity muscles. These recordings are obtained while the monkey performs tasks involving reaching, grasping, and manipulating four different objects positioned at eight different locations. The objects include various shapes such as cylinders, buttons, spheres, etc., and the monkey is trained to perform specific manipulation actions for each object type. Trials begin with the monkey interacting with a central cylinder, followed by cues indicating which peripheral object to manipulate. Successful trials involve the monkey completing the required interactions with the cued object within specified time frames. This data is provided by Adam Rouse at University of Kansas.
Why M1?
Successfully working with this dataset requires high-accuracy and consistent EMG decoding performance. EMG decoding is particularly challenging as it requires a model to forecast multiple output variables (here, 16 muscles). Moreover, EMG decoding bears relevance to BCI developments aiming to apply functional electrical stimulation (FES) for inducing movement in a user's limb. However, EMG data is costly and difficult to collect as it requires an invasive implant in the muscles, and many BCI users may not have muscular control that can yield high fidelity EMG recordings.
As a first step, developing methods to improve the stability of EMG decoding would limit the demand to collect new EMG calibration data, thus greatly improving the practical viability of EMG-based neuroprosthetic motor control. Future endeavors may also aim to develop methods for cross-participant transfer to increase the amount of high-quality EMG data available for BCI applications.
M2

M2 is a dataset of a monkey moving two finger groups to reach targets cued on a screen. The dataset has neural activity from one Utah array, and finger position data as recorded by the state of the finger manipulandum that the monkey's fingers were enclosed in. The monkey has been trained to do this task and the data is collected in the context of a BCI experiment for finger control. The data is provided by the Chestek lab at University of Michigan.
Why M2?
Finger control is a critical aspect of dexterous hand function and is a key target for neuroprosthetic control. M2's continuous finger behavior is a non-prehensile behavior, distinguishing it from the other major class of prehensile, grasping behavior (as in M1). Further, recent work, as in Nason et al., 21, where this data was collected and Shah et al., 23, are identifying linear compositionality in the encoding of different finger behaviors in motor cortex. However, this structure has unclear implications for the stability of decoding finger movements for BCI control.
B1
B1 contains data from zebra finch songbirds vocalizing in a naturalistic setting. The dataset consists of neural activity recorded from premotor and motor brain regions, collected with Neuropixels 1.0 probes, as well as simultaneous audio recordings of the vocalizations. The songs are annotated with template-matching software to identify repeated syllables and motifs. This data is provided by the Translational Neuroengineering Lab and the Gentner Lab, both at UC San Diego.
Why B1?
B1 is unique in that it is the only dataset in the FALCON benchmark recorded using Neuropixels probes, which may exhibit nonstationarities of different nature than the Utah and microelectrode arrays used in the other datasets. Also, because songbird vocalization parallels human speech (Anumanchipalli et al., 19), B1 enables evaluation of iBCI decoding methods that may be adapted for human speech decoding.